Recent advances in immunotherapy have highlighted that in multiple myeloma (MM), besides risk-determining copy number alterations (CNAs) there is a need for investigation of CNAs in CAR-T cell therapy-related targets (e.g. BCMA, GPRC5D, FcRH5). DigitalMLPA technology has been successfully used for characterization of recurrent CNAs on chromosome arms 1p, 1q, 13q and 17p in newly diagnosed and relapsed MM cases, as well as in cancer cell lines of MM origin. Adding probes aimed at CAR-T cell therapy-related target genes complements the already included predictive target probes ( IKZF1/3, IRF4, MYC, RPL5, SLAMF7 and BRAF V600E point mutation) in SALSA® digitalMLPA™ Probemix D006 Multiple Myeloma. In addition, due to a large overlap of affected genomic regions between hematologic neoplasms, we aimed to demonstrate usefulness of this multiplex assay not only on cell lines of MM origin, but also of myeloid and lymphoid lineages.
Methods. D006 Multiple Myeloma digitalMLPA probemix contains (i) probes targeting chromosomal arms with recurrent CNAs, including 1p (33 probes), 1q (29 probes), 13q (23 probes), 17p (20 probes); (ii) probes for promising predictive targets such as BCMA, CRBN, GPRC5D, FcRH5, IKZF1/3, IRF4, MYC, RPL5, SLAMF7 - 2-4 probes per gene, and BRAF V600E mutation-specific probe; (iii) 278 probes for subtelomeric, pericentromeric and middle regions of chromosomal arms for all chromosomes; and (iv) control probes for quality control and troubleshooting. DigitalMLPA reactions were performed with 20-40 ng DNA from 29 cancer cell lines of lymphoid and myeloid lineage, 16 of which were of MM origin. Commercial blood-derived genomic DNA samples from healthy individuals were used as references for data normalization using data analysis software Coffalyser digitalMLPA™.
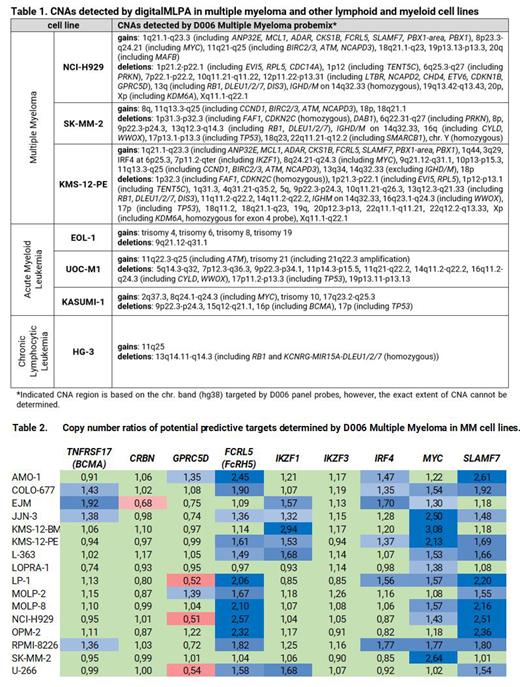
Results. Losses of 1p were among the most frequent CNAs in MM cell lines (n=13) and were mainly local (1p21.1-p22.1), including homozygous deletion of CDKN2C in MM cell lines co-occurring with TENT5C subclonal loss in 2 of those. 13q loss (encompassing predominantly the whole arm) was detected in 13 MM cell lines. 1q and 17p losses were present in 12 MM cell lines. One MM cell line showed loss of BCMA (LOPRA-1) and one of CRBN (EJM), 6 had loss of GPRC5D, 9 had MYC gain. None of the MM cell lines carried the BRAF V600E mutation. In the EOL-1 myeloid lineage cell line, trisomy of chromosomes 4, 6, 8 and 19 were correctly identified by increased ratios for all probes covering these chromosomes: 24, 27, 20 and 11 probes respectively; and in the HG-3 lymphoid lineage cell line 11q25 gain and 13q loss was detected. Examples of CNAs detected by D006 Multiple Myeloma probemix are described in Table 1. Copy number ratios of predictive target genes in MM cell lines are shown in Table 2. Ratios between 0.7-1.3 (green cells) indicate normal copy number, ratios below 0.7-1.3 range indicate copy number loss (red cells) and higher ratios indicate gains (blue cells). CNAs detected by digitalMLPA were highly concordant with those reported in public databases.
Conclusions. digitalMLPA technology is well suited for multiplex CNA detection of routinely analyzed genomic regions in MM and provides a unique opportunity to research molecular genetic markers of emerging significance in MM in the same reaction. In addition, this MM assay also gives the possibility to analyze gross CNAs in other hematopoietic neoplasms with overlapping affected regions. Input DNA requirements that are a fraction of current WGS, simple protocol and possibility of combining of digitalMLPA libraries with other NGS libraries make D006 Multiple Myeloma digitalMLPA probemix a reliable, cost-effective and robust method to detect well-established and emerging CNAs.
Disclosures
Atanesyan:MRC Holland: Current Employment. Enright:MRC Holland: Current Employment. Kaiser:GSK: Consultancy; Celgene/BMS: Consultancy, Honoraria, Research Funding; Janssen: Consultancy, Honoraria; Regeneron: Consultancy; Takeda: Honoraria; Seagen: Consultancy; Karyopharm: Consultancy; Pfizer: Consultancy. Savola:MRC Holland: Current Employment.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal